Scientists have developed a powerful AI model that can create instructions for making brand-new proteins, going beyond what exists in nature. Since proteins are the building blocks of life, this breakthrough could lead to exciting new possibilities in medicine, technology, and science.
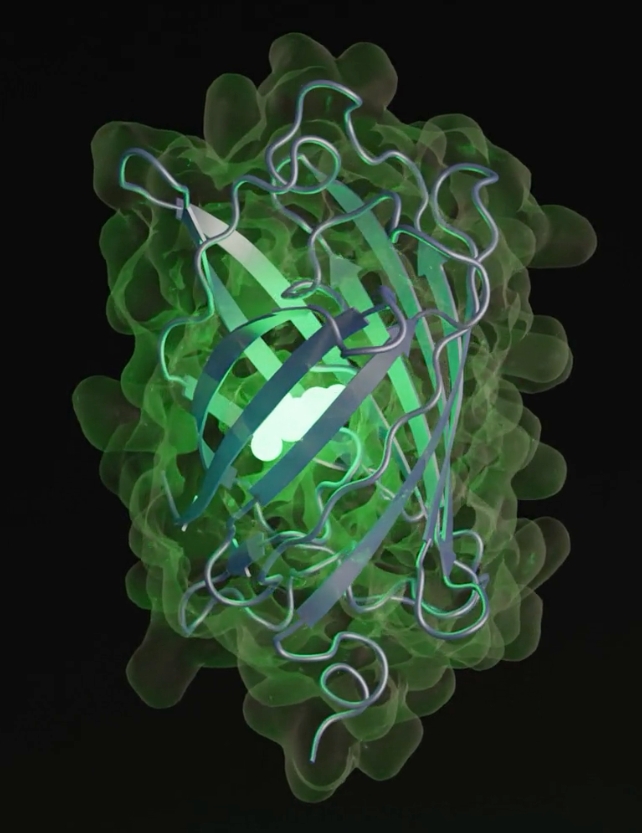
Scientists in the US have used an advanced AI model, EvolutionaryScale Model 3 (ESM3), to create a new green fluorescent protein called esmGFP. This protein is only 58% similar to its closest natural relative, tagRFP.
According to researchers, this is like compressing 500 million years of evolution into -driven design. This breakthrough could lead to custom-made proteins for specific purposes or help discover new functions in existing proteins.
We show that large language models trained on evolutionary data can create new, working proteins that are very different from existing ones.
Scientists estimate that AI has processed the equivalent of 500 million years of evolution. This breakthrough could help create custom-designed proteins for specific purposes or discover new functions in existing proteins.
esmGFP is special because it works—it glows, just like tagRFP. Fluorescent proteins make some ocean creatures glow and are very useful in medicine and biotechnology.
By analyzing large amounts of data, the AI model learns what helps proteins work and what doesn’t—just like how ChatGPT can write a rhyming poem after reading many poems written by people.
More than three billion years of evolution have shaped the design of life, storing biological information within natural proteins,” wrote researchers led by Thomas Hayes, founder of EvolutionaryScale in New York, in their published paper.
We show that large language models trained on evolutionary data can create new
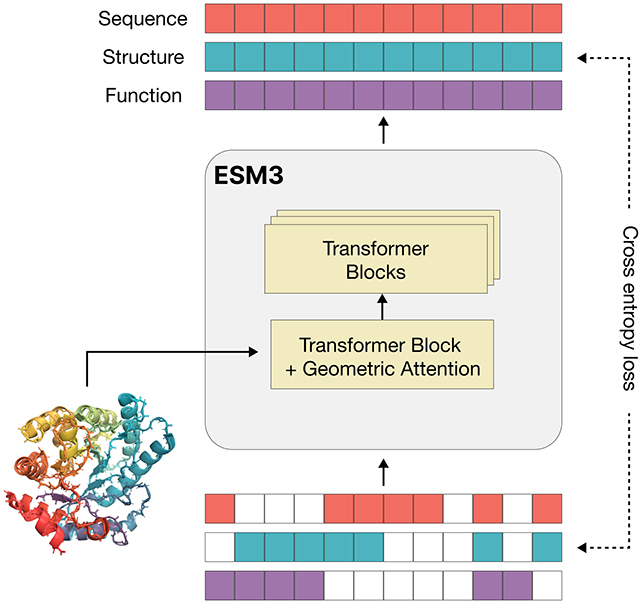
ESM3 was trained using a vast amount of data, including 3.15 billion protein sequences (the order of amino acids in a protein), 236 million protein structures (their 3D shapes), and 539 million protein annotations (descriptive labels).
can learn from huge amounts of data to find patterns in protein building and function—just like ChatGPT learns to write poems by reading many human-written ones.
What makes esmGFP special is that it actually works! It glows like its relative, tagRFP. Fluorescent proteins help some ocean creatures shine and are important tools in medicine and biotechnology.
We chose fluorescence because it is hard to achieve, easy to measure, and one of the most beautiful natural mechanisms,” the team writes.
AI reduces guesswork in protein synthesis and helps discover completely new proteins beyond what we already know.
Proteins exist in a connected space where each one is linked to others by small genetic changes. Evolution moves through this space, changing proteins step by step while keeping their function intact.
helps remove much of the guesswork in protein synthesis and allows scientists to explore entirely new proteins beyond what we currently know.
Researchers explain that proteins exist in a connected space, where each one is just one small change (mutation) away from another. Evolution follows a network of these connections, linking proteins through possible changes over time.
For evolution to work, each protein must change in a way that keeps the overall system functioning. A language model can help recognize and predict proteins within this network, making the process more efficient.
A language model helps understand proteins in this space. The model, ESM3, can design new proteins, but these still need to be tested and developed. Scientists believe that soon, could help create proteins for medicines, biomaterials, and more—just by using smart prompts.
Also Read….. Thandel Box Office Day 1 Collection: Naga Chaitanya and Sai Pallavi’s Romantic Thriller Starts Strong















 Categories
Categories